Deep Learning from Scratch to GPU - 3 - Fully Connected Inference Layers
February 14, 2019
Please share this post in your communities. Without your help, it will stay burried under tons of corporate-pushed, AI and blog farm generated slop, and very few people will know that this exists.
These books fund my work! Please check them out.
If you haven't yet, read my introduction to this series in Deep Learning in Clojure from Scratch to GPU - Part 0 - Why Bother?.
The previous article, Part 2, is here: Bias and Activation Function.
To run the code, you need a Clojure project with Neanderthal () included as a dependency. If you're in a hurry, you can clone Neanderthal Hello World project.
Don't forget to read at least some introduction from Neural Networks and Deep Learning, start up the REPL from your favorite Clojure development environment, and let's continue with the tutorial.
(require '[uncomplicate.commons.core :refer [with-release let-release Releaseable release]] '[uncomplicate.neanderthal [native :refer [dv dge]] [core :refer [mv! axpy! scal! transfer!]] [vect-math :refer [tanh! linear-frac!]]] '[criterium.core :refer [quick-bench]])
The updated network diagram
We'll update the neural network diagram, to reflect the recently included biases and activation functions.
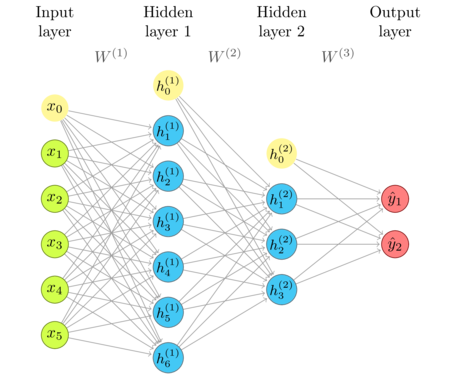
In each layer, bias is shown as an additional, "zero"-indexed, node, which, connected to nodes in the next layer, gives the bias vector. I haven't explicitly shown activation functions to avoid clutter. Consider that each node has activation at the output. If a layer didn't have any activation whatsoever, it would have been redundant (see composition of transformations).
The Layer type
We need a structure that keeps the account of the weights, biases and output of each layer.
It should manage the life-cycle and release that memory space when appropriate.
That structure can implement Clojure's IFn interface, so we can invoke it as any regular
Clojure function.
(import 'clojure.lang.IFn)
We will create FullyConnectedInference as a Clojure type, which gets compiled into a Java class.
FullyConnectedInference implements the release method of the Releaseable protocol,
and the invoke method of IFn. We would also like to access and see the weights and bias values
so we create new protocol, Parameters, with weight and bias methods.
(defprotocol Parameters (weights [this]) (bias [this])) (deftype FullyConnectedInference [w b h activ-fn] Releaseable (release [_] (release w) (release b) (release h)) Parameters (weights [this] w) (bias [this] b) IFn (invoke [_ x] (activ-fn b (mv! w x h))))
Constructor function
At the time of creation, each FullyConnectedInference need to be supplied with an activation function,
the matrix for weights, and vectors for bias and output that have matching dimensions. Of course,
we will automate that process.
(defn fully-connected [activ-fn in-dim out-dim] (let-release [w (dge out-dim in-dim) bias (dv out-dim) h (dv out-dim)] (->FullyConnectedInference w bias h activ-fn)))
Now a simple call to the fully-connected function that specifies the activation function and
the dimensions of input dimension (number of neurons in the previous layer) and output dimension
(the number of neurons in this layer) will create and properly initialize it.
let-release is a variant of let that releases its bindings (w, bias, and/or h) if
anything goes wrong and an exception gets thrown in its scope.
with-release, on the other hand, releases the bindings in all cases.
Activation functions
We can use a few matching functions that we discussed in the previous article.
(defn activ-sigmoid! [bias x] (axpy! -1.0 bias x) (linear-frac! 0.5 (tanh! (scal! 0.5 x)) 0.5))
(defn activ-tanh! [bias x] (tanh! (axpy! -1.0 bias x)) )
Using the fully-connected layer
Now we are ready to re-create the existing example in a more convenient form. Note that we no longer
have to worry about creating the matching structures of a layer; it happens automatically when
we create each layer. I use the transfer! function to set up the values of weights and bias,
but typically these values will already be there after the training (learning) of the network. Here
we are using the same "random" numbers as before as a temporary testing crutch.
(with-release [x (dv 0.3 0.9) layer-1 (fully-connected activ-sigmoid! 2 4)] (transfer! [0.3 0.1 0.0 0.0 0.6 2.0 3.7 1.0] (weights layer-1)) (transfer! (dv 0.7 0.2 1.1 2) (bias layer-1)) (println (layer-1 x)))
#RealBlockVector[double, n:4, offset: 0, stride:1] [ 0.48 0.84 0.90 0.25 ]
The output is the same as before, as we expected.
Multiple hidden layers
Note that the layer is a transfer function that, given the input, computes and returns the output. Like with other Clojure functions, the output value can be an input to the next layer function.
(with-release [x (dv 0.3 0.9) layer-1 (fully-connected activ-tanh! 2 4) layer-2 (fully-connected activ-sigmoid! 4 1)] (transfer! [0.3 0.1 0.9 0.0 0.6 2.0 3.7 1.0] (weights layer-1)) (transfer! [0.7 0.2 1.1 2] (bias layer-1)) (transfer! [0.75 0.15 0.22 0.33] (weights layer-2)) (transfer! [0.3] (bias layer-2)) (println (layer-2 (layer-1 x))))
#RealBlockVector[double, n:1, offset: 0, stride:1] [ 0.44 ]
If we don't count the transfer! calls, we have 3 lines of code that describe the whole network:
two lines to create the layers, and one line for the nested call of layers as functions. It is already
concise, and we will still improve it in the upcoming articles.
Micro benchmark
With rather small networks consisting of a few layers with few neurons each, any implementation will be fast. Let's create a network with a larger number of neurons, to get a feel of how fast we can expect these things to run.
Here is a network with input dimension of 10000. The exact numbers are not important, since the example is superficial, but you can imagine that 10000 represents an image that has 400 × 250 pixels. I've put 5000 neurons in the first layer, 1000 in the second layer, and 10 at the output. Just some random large-ish dimensions. Imagine 10 categories at the output.
(with-release [x (dv 10000) layer-1 (fully-connected activ-tanh! 10000 5000) layer-2 (fully-connected activ-sigmoid! 5000 1000) layer-3 (fully-connected activ-sigmoid! 1000 10)] ;; Call it like this: ;; (quick-bench (layer-3 (layer-2 (layer-1 x)))) (layer-3 (layer-2 (layer-1 x))))
Evaluation count : 36 in 6 samples of 6 calls.
Execution time mean : 18.320566 ms
Execution time std-deviation : 1.108914 ms
Execution time lower quantile : 17.117256 ms ( 2.5%)
Execution time upper quantile : 19.847416 ms (97.5%)
Overhead used : 7.384194 ns
On my 5 year old CPU (i7-4790k), one inference through this network takes 18 milliseconds. If you have another neural networks library at hand, you could construct the same network (virtually any NN software should support this basic structure) and compare the timings.
The next article
So far so good. 18 ms should not be slow. But what if we have lots of data to process? Does that mean that if I have 10 thousand inputs, I'd need to make a loop that invokes this inference 10 thousand times with different input, and wait 3 minutes for the results?
Is the way to speed that up putting it on the GPU? We'll do that later, but there is something that we can do even on the CPU to improve this! That's the topic of the next article, Increasing Performance with Batch Processing!
Thank you
Clojurists Together financially supported writing this series. Big thanks to all Clojurians who contribute, and thank you for reading and discussing this series.

